-
Notifications
You must be signed in to change notification settings - Fork 3k
Float point embeddings
jingkl edited this page Oct 22, 2021
·
3 revisions
| Similarity Metrics | Index Types |
|---|---|
|
|
Essentially, Euclidean distance measures the length of a segment that connects 2 points.
The formula for Euclidean distance is as follows:
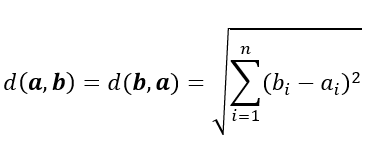
where a = (a1, a2,..., an) and b = (b1, b2,..., bn) are two points in n-dimensional Euclidean space
It's the most commonly used distance metric and is very useful when the data is continuous.
The IP distance between two embeddings are defined as follows:

Where A and B are embeddings, ||A|| and ||B|| are the norms of A and B.
IP is more useful if you are more interested in measuring the orientation but not the magnitude of the vectors.
If you use IP to calculate embeddings similarities, you must normalize your embeddings. After normalization, the inner product equals cosine similarity.
Suppose X' is normalized from embedding X:
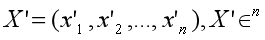
The correlation between the two embeddings is as follows:
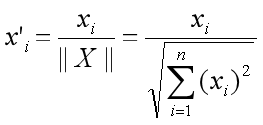
For example:
In python
from pymilvus import connections, FieldSchema, CollectionSchema, DataType, Collection
# create a collection
collection_name = "milvus_test"
default_fields = [
FieldSchema(name="id", dtype=DataType.INT64, is_primary=True, auto_id=True),
FieldSchema(name="vector", dtype=DataType.FLOAT_VECTOR, dim=d)
]default_schema = CollectionSchema(fields=default_fields, description="test collection")
print(f"\nCreate collection...")
collection = Collection(name= collection_name, schema=default_schema)
# insert data
import random
vectors = [[random.random() for _ in range(8)] for _ in range(10)]
entities = [vectors]
mr = collection.insert(entities)
print(collection.num_entities)
# create index
collection.create_index(field_name=field_name,
index_params={'index_type': 'IVF_FLAT',
'metric_type': 'L2',
'params': {
"M": 16, # int. 4~64
"efConstruction": 40 # int. 8~512
}})
collection.load()
# search
top_k = 10
search_params = {"metric_type": "L2", "params": {"nprobe": 10}}
results = collection.search(vectors[:5], anns_field="vector", param=search_params,limit=top_k)
# show results
for result in results:
print(result.ids)
print(result.distance)